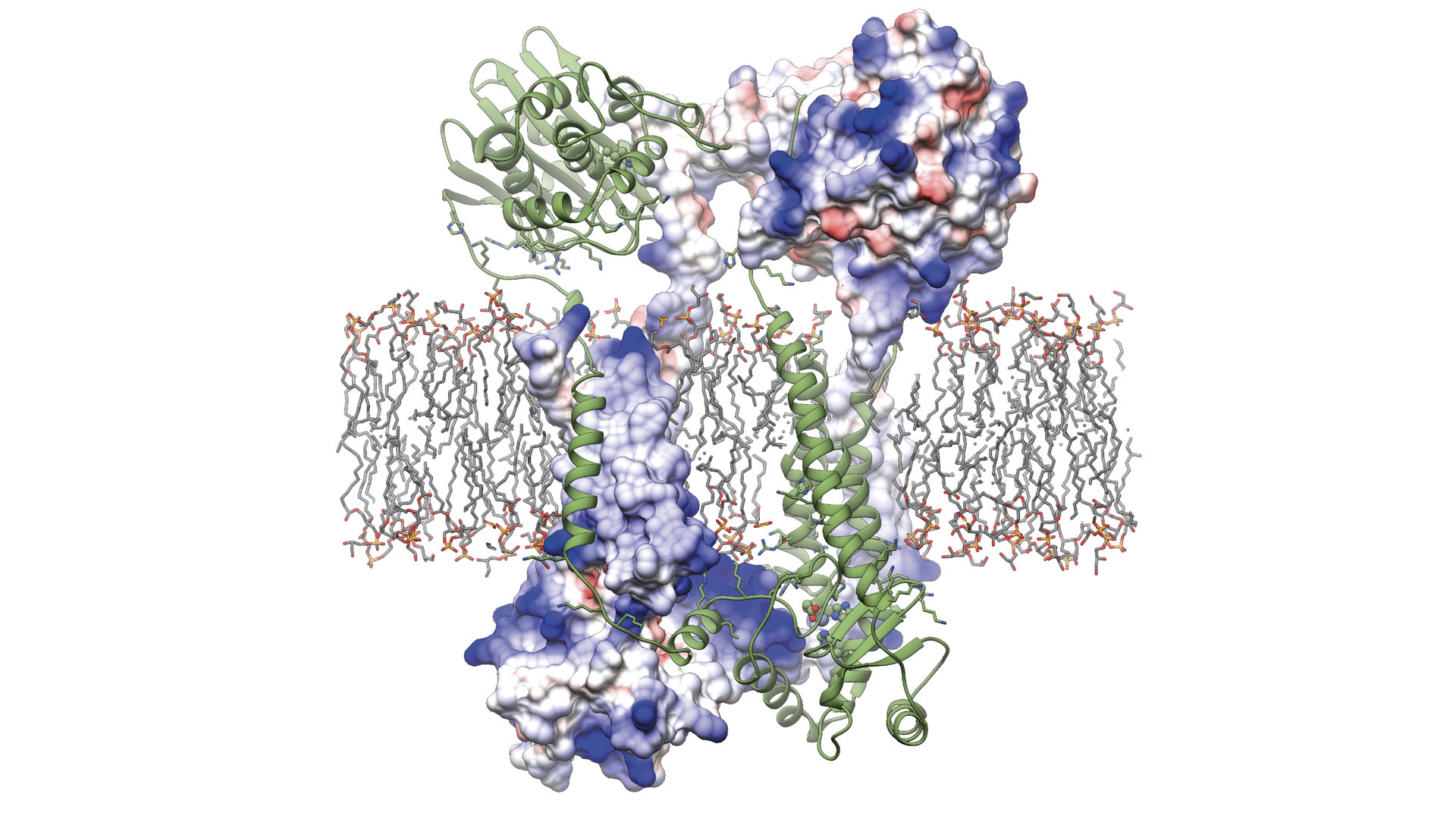
UBC study reveals how the protein Blar1 (pictured above) controls antibiotic resistance in Staphylococcus aureus.
A recent study led by UBC researchers is shedding light on how one of the world’s most notorious superbugs, Staphylococcus aureus (S. aureus), develops resistance to antibiotics.

Dr. Natalie Strynadka
The findings, published in Nature, provide critical insights that will help drive the development of more effective therapeutics to combat the growing global threat of drug-resistant bacterial infections.
“The rise of antibiotic-resistant bacteria has rendered many life-saving drugs ineffective, jeopardizing our ability to combat infectious disease,” says the study’s senior author Dr. Natalie Strynadka, a distinguished professor of biochemistry and molecular biology at UBC, and Tier 1 Canada Research Chair in structure-guided antibiotic discovery. “Having a clearer understanding of how resistance develops is absolutely critical for facilitating a new cocktail of therapies that preserve the effectiveness of existing antibiotics.”
S. aureus is a bacterium commonly found on the skin of healthy individuals. However, it can cause potentially life-threatening infections when it enters the body through a wound or other means.
Of particular concern is the ability of S. aureus to adapt and develop resistance to beta-lactam antibiotics, like penicillin, that are one of the most commonly prescribed drugs to treat infections globally. New strains including methicillin-resistant S. aureus, or MRSA, have caused significant outbreaks in healthcare and community settings globally, leading the World Health Organization to include S. aureus on their list of priority pathogens that pose the greatest threat to human health.

Members of the High Resolution Macromolecular Cryo-Electron Microscopy lab at UBC (Left to right: Natalie Strynadka, Marija Vuckovic, Liam Worrall, Claire Atkinson, Joeseph Felt.)
A microscopic master switch
For the study, the research team used cryo-electron microscopy (cryo-EM) to determine the molecular structure of a key protein, BlaR1, that exists in the cell membrane of S. aureus and is known to play a key role in resistance to beta-lactam antibiotics.
Cryo-EM is a powerful imaging technique that uses a beam of electrons to map the three-dimensional structure of biological macromolecules. Until now, the structure of BlaR1 and the mechanisms by which it enables antibiotic resistance has eluded scientists worldwide. The UBC team overcame previous barriers by using novel techniques to purify and isolate the protein, work led by PhD student Andrew Alexander and research technologist Marija Vuckovic.
“The question now becomes, how can we cut the circuit and turn off that resistance in one fell swoop by targeting BlaR1.”
Dr. Natalie Strynadka
“This is the first study to present the full-length structure of this important protein,” said Dr. Liam Worrall, Research Associate, who led the data processing and structure analysis in the study with assistance from PhD student Jinhong Hu. “With cryo-EM, we’re able to visualize how S. aureus detects and reacts to the presence of beta-lactam antibiotics, providing atomic level understanding of the first critical steps of cellular events that take place to enable antibiotic resistance.”
The three-dimensional structure of BlaR1 was determined using the open access High Resolution Macromolecular Cryo-Electron Microscopy facility in the Life Sciences Institute at UBC.

Liam Worrall
The study’s findings reveal that the BlaR1 protein changes shape when it comes into contact with beta-lactam antibiotics. This sends signals across the cell membrane, triggering a series of steps that instruct the bacteria to produce enzymes and proteins that give it antibiotic-fighting superpowers.
For Dr. Strynadka, it’s an important breakthrough that will allow scientists to fight back against anti-microbial resistance.
“BlaR1 is like a master switch controlling broad-spectrum beta-lactam antibiotic resistance mechanisms in S. aureus. The question now becomes, how can we cut the circuit and turn off that resistance in one fell swoop by targeting BlaR1,” said Dr. Strynadka.
The researchers are now turning their attention to identifying ways of intervening in this cellular pathway, which could take the form of small molecule drugs or vaccines. The potential of such drugs could extend beyond S. aureus, as similar signaling systems have been identified in other high-priority pathogens such as Clostridium difficile and Mycobacterium tuberculosis.
With more than a million people a year estimated to die from antibiotic-resistant infections, and a dwindling number of new antibiotics being discovered, it’s urgent work.
“We have fewer and fewer tools at our disposal to fight these superbugs, threatening over a century of medical progress,” said Dr. Strynadka. “This study opens up new avenues for understanding and targeting antibiotic resistance in a range of priority pathogens, ensuring the preservation of the beta-lactam antibiotics for combating infectious diseases.”